BayesSEIR
Overview
$\texttt{BayesSEIR}$ is an R package developed to fit fully Bayesian discrete-time stochastic SEIR models of epidemic spread. The package offers modelers three methods to describe the infectious period: exponentially distributed, gamma or Weibully distributed using the path-specific approach of Porter and Oleson (2013), and the infectious duration-dependent transmissibility proposed by the package authors.
Model Details
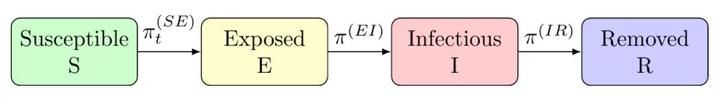
The SEIR model includes four compartments, Susceptible, Exposed, Infectious, and Removed/Recovered. BayesSEIR uses the chain binomial structure as presented in Lekone and Finkenstädt (2006). The model assumes a closed population of size $N$. Let $t = 1, …, \tau$ indicate discrete calendar time since the epidemic began and $S_t$, $E_t$, $I_t$, and $R_t$ denote the number of individuals in the susceptible, exposed, infectious, and removed compartment in the time interval $(t, t+1]$, respectively. Furthermore, define $E^\ast_t$, $I^\ast_{t}$, and $R^\ast_{t}$ to represent the number of individuals that transition into the indicated compartment in this interval.
The counts of newly exposed, infectious, and removed individuals is modeled using a chain binomial structure:
$$E^\ast_t \sim Bin\Big(S_t, \pi_t^{(SE)}\Big)$$
$$I^\ast_{t} \sim Bin\Big(E_{t}, \pi^{(EI)} \Big)$$
$$R^\ast_{t} \sim Bin\Big(I_{t}, \pi^{(IR)} \Big)$$
The transition probabilities, $\pi_t^{(\text{SE})}$, $\pi^{(\text{EI})}$, and $\pi^{(\text{IR})}$ are of primary interest. $\texttt{BayesSEIR}$ assumes the transmission probability is written as
$$\pi_t^{(\text{SE})} = 1 - \exp \Big(-e^{\theta_t} \frac{I_t}{N}\Big)$$
where $\theta_t = X_t' \boldsymbol{\beta}$, $X_t$ is a $1 \times p$ row-vector, and $\boldsymbol{\beta}$ is a $p \times 1$ vector used to describe the intensity process.
$\texttt{BayesSEIR}$ uses an exponential distribution to describe the time spent in the latent period, which translates to a probability written as
$$\pi^{(\text{EI})} = 1 - \exp (- \rho_E)$$
where $\rho_E$ is the rate parameter, such that $1/\rho_E$ gives the mean duration of the latent period.
$\texttt{BayesSEIR}$ offers three possible methods for specifying the infectious period:
-
Exponentially distributed: $$\pi^{(\text{IR})} = 1 - \exp (- \rho_I)$$ where $\rho_I$ is the rate parameter, such that $1/\rho_I$ gives the mean duration of the infectious period.
-
Path-specific: $$\pi^{(\text{IR})} = P(W \leq w + 1 | W > w)$$ where $W$ can follow an exponential, gamma, or Weibull distribution.
-
Infectious duration-dependent (IDD) transmission: $$\pi_t^{(\text{SE})} = 1 - \exp \Big(-e^{\theta_t} \frac{ \sum_{w = 1}^{T_I} f(w) I_{wt} }{N} \Big)$$ where the duration of the infectious period is fixed at $T_I$ days and $f(w) > 0$ describes a curve of transmissibility over each individual’s infectious period. BayesSEIR offers four functions that can be used to specify $f(w)$, the gamma PDF, the log normal PDF, a logistic decay function, and a function derived from splines.
Examples
For some examples of the package in action, check out the package vignettes:
Simulating Epidemics and Computing R0
References
Lekone, P. E., & Finkenstädt, B. F. (2006). Statistical inference in a stochastic epidemic SEIR model with control intervention: Ebola as a case study. Biometrics. DOI: 10.1111/j.1541-0420.2006.00609.x
Porter, A. T., & Oleson, J. J. (2013). A path-specific SEIR model for use with general latent and infectious time distributions. Biometrics. DOI: 10.1111/j.1541-0420.2012.01809.x
Ward, C., Brown, G. D., & Oleson, J. J. (2022) Incorporating Infectious Duration-Dependent Transmission into Bayesian Epidemic Models. Biometrical Journal. DOI: 10.1002/bimj.202100401